
PharmAI at BIO-Europe Digital 2020
September 18, 2020
PharmAI at BIO-Europe Spring Digital 2021
March 1, 2021Running Molecular Dynamics with Simulated Annealing in OpenMM
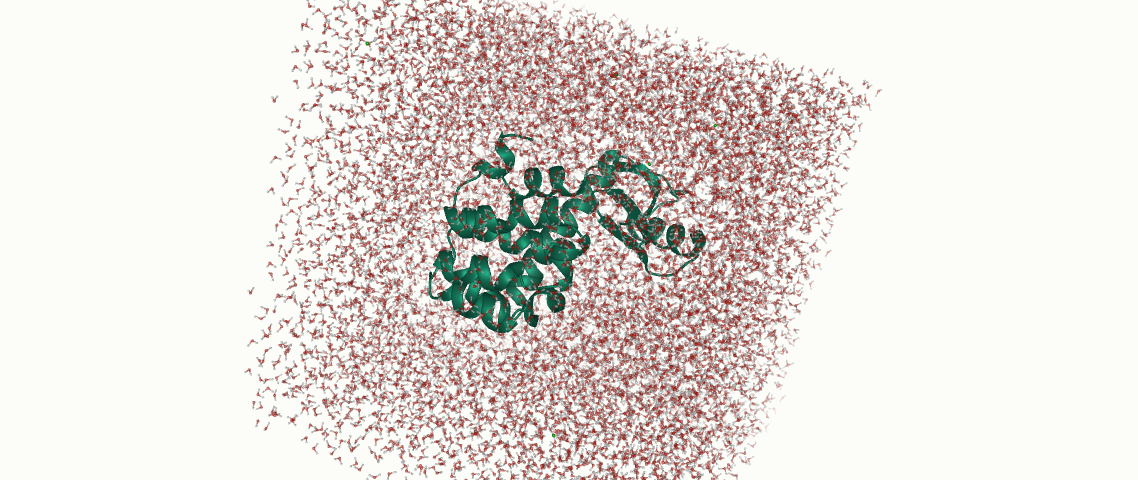
Molecular dynamics is a fantastic tool to understand the behavior of a chemical system over time. In this blog post, we share an easy recipe for a simulation with OpenMM that involves a part of simulated annealing.
Simulated Annealing
Simulated annealing in the context of molecular dynamics refers to the controlled heating and cooling of the system in order to overcome energetic barriers and find energetically favorable minima much faster. This process can be repeated multiple times over the course of simulation.

Setting-Up Simulated Annealing with OpenMM
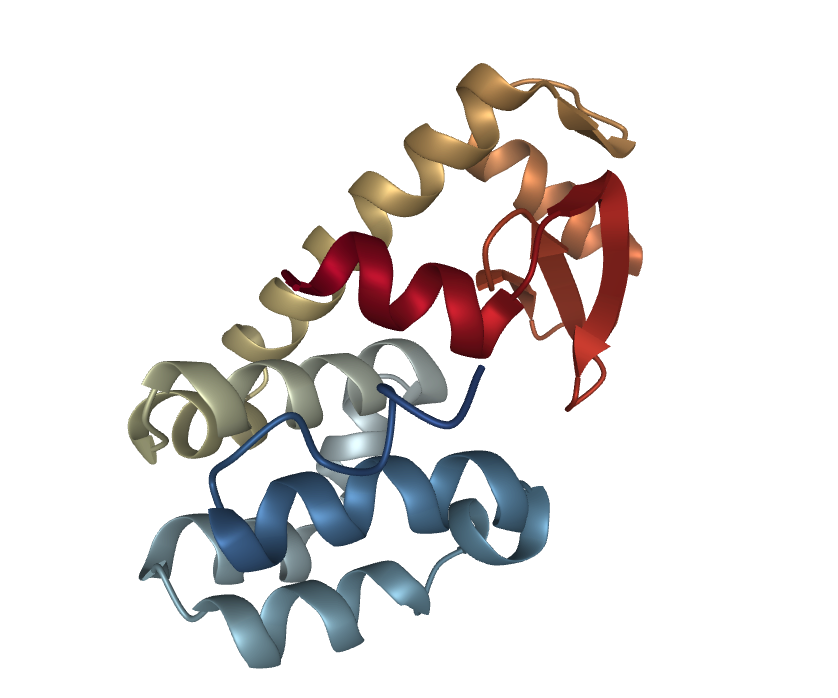
In the following we will use T4 Lysozyme as a toy example in our simulation. We want to simulate the annealing with the following temperatures:
| starting temerature | 0 K |
| normal temperature | 300 K (26.85 °C) |
| highest temperature | 315 K |
Creating the System
We use OpenMM Setup to create the system, solvate the protein, and to fix potential errors in our PDB files. It is highly recommended to prepare your system with this tool, as it will avoid potential errors later.

The protein was solvated in a 10 Å water box, hydrogens and neutralizing ions were added:
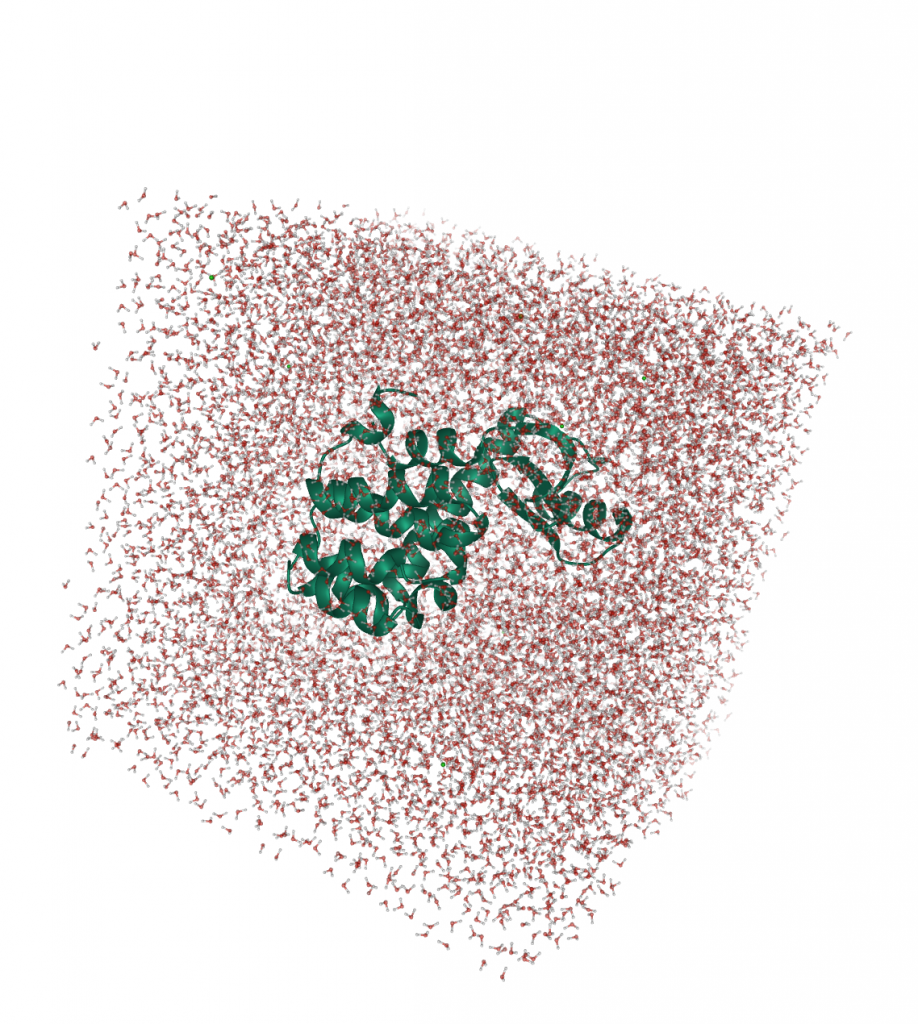
If you want to reproduce the simulation, you can download the pre-processed PDB file here and the simulation protocol here.
A snippet of the most important part of the simulation is shown below:
...
# Phase I
print(f'initial heating of system')
for temp in np.arange(start_temperature.value_in_unit(kelvin),end_temperature.value_in_unit(kelvin),temperature_step.value_in_unit(kelvin)):
if temp % 1 == 0:
print(f'current temperature: {temp}')
integrator.setTemperature(temp*kelvin)
barostat.setDefaultTemperature(temp*kelvin)
simulation.step(heating_interval)
# Phase II
print(f'simulating')
simulation.step(steps)
# Phase III
print(f'heating system to top temperature')
for temp in np.arange(end_temperature.value_in_unit(kelvin),top_temperature.value_in_unit(kelvin),top_temperature_step.value_in_unit(kelvin)):
print(f'current temperature: {temp}')
integrator.setTemperature(temp*kelvin)
barostat.setDefaultTemperature(temp*kelvin)
simulation.step(heating_interval)
# Phase IV
print(f'holding top temperature')
simulation.step(100000)
# Phase V
print(f'cooling system to end temperature')
for temp in np.arange(top_temperature.value_in_unit(kelvin),end_temperature.value_in_unit(kelvin),-top_temperature_step.value_in_unit(kelvin)):
print(f'current temperature: {temp}')
integrator.setTemperature(temp*kelvin)
barostat.setDefaultTemperature(temp*kelvin)
simulation.step(heating_interval)
# Phase VI
print(f'final equilibration')
simulation.step(300000)
...
